- project instruction
Figure 1. Chemical structure of compound 1.
2. Calculation method
The X-ray crystal structure of PAP1 (PDB number: 4RV6, resolution: 3.19 Ã…) was downloaded from the RCSB Protein Data Bank (http://) with the first conformation as the acceptor structure.
[1]. The three-dimensional structure of Compound 1 was established using UCSF Chimera software and energy optimization was performed.
[2,3]. Add hydrogen atoms using the Dock Prep module and add AMBER ff14SB force field and AM1-BCC charge, respectively. The molecular surface of the receptor was generated using a DMS tool in Chimera with a probe with a radius of 1.4 Ã….
[4,5]. X-ray crystal structure shows a reasonable binding site, for which the sphgen module is used to generate a spherical collection (Spheres) around the active site, and the Grid module is used to generate the Grid file. The file is used for Grid-based energy scoring evaluation. The DOCK6.7 program was used for semi-flexible docking to generate 10,000 different conformational orientations and to obtain electrostatic and van der Waals interactions between the ligand molecules and the binding sites, and the Grid scores were calculated therefrom. By cluster analysis (RMSD threshold 2.0Ã…), the best scoring is obtained.
[6]. Finally, PyMOL is used to generate images.
3. Calculation results
A. Combining conformation scores
The binding pattern of Compound 1 in PARP1 was predicted using the DOCK6.7 program, retaining up to 20 binding conformations. The calculation results show that the binding sites have multiple docking conformations, and the scoring conditions are as follows (Table 1). The second docking conformation is selected according to the scoring and combining modes for combined mode analysis.
Table 1. Docking scores for Compound 1 and Receptor PARP1 (unit: kcal/mol)
| Compound | Pose | Grid Score | Grid_vdw | Grid_es | Int_energy |
| AG14361 | 1 | -58.413887 | -57.743397 | -0.670492 | 6.865056 |
| 2 | -55.357056 | -53.782494 | -1.574563 | 5.41468 | |
| 3 | -55.327587 | -55.685509 | 0.35792 | 5.660656 |
B. Combined mode analysis
The amide carbonyl oxygen atom on the seven-membered ring of compound 1 forms a hydrogen bond interaction with the amino acid residues Ser904 and His862; at the same time, the amide nitrogen atom forms a hydrogen bond interaction with Gly863 of 3.33 Ã…. This anchors the direction of the compound and provides a certain electrostatic contribution (Grid_es = -1.574563 kcal/mol).
P-type π-π stacking between the two rings of benzimidazole and Tyr90, the center distance of the aromatic ring is 4.21 Å and 4.93 Å, respectively; T-type π-π stacking between the side chain benzene ring and Tyr896, Fang The center distance of the ring is 5.47 Å. At the same time, the compound also forms a hydrophobic interaction with the residues Tyr889, Tyr896, Tyr907 and Glu998. The hydrophobic interaction and π-π stacking provide a strong van der Waals force for the compound (Grid_vdw = -53.78 kcal/mol).
In summary, the interaction of Compound 1 with protein PARP1 is dominated by π-π stacking and hydrophobic interaction, and the binding orientation is locked by hydrogen bonding.
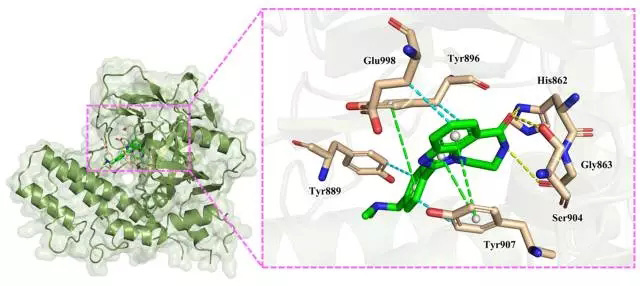
Figure 2. Binding pattern of compound 1 to protein (for a detailed description, see Legend)
references:
[1]. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE. Ucsf chimera–a visualization system for exploratory research and analysis. J Comput Chem, 25(13): 1605–12 , 2004.
[2]. Araz Jakalian, Bruce L. Bush, David B. Jack, and Christopher I. Bayly. Fast, efficient generation of high-quality atomic charges. am1-bcc model: I. method. Journal of Computational Chemistry, 21 (2 ): 132–146, January 2000.
[3].Araz Jakalian, David B.Jack, and Christopher I. Bayly. Fast, efficient generation of high-quality atomic charges. am1-bcc model: I.parameterization and validation Journal of Computational
Chemistry, 23(16): 1623–1641, December 2002.
[4].P.Therese Lang, Scott R.Brozell, Sudipto Mukherjee, Eric F.Pettersen, Elaine C.Meng, Veena Thomas, Robert C.Rizzo, David A.Case, Thomas L.James James, and Irwin D. Kuntz.Dock 6: Combining techniques to model rna-small molecule complexes. RNA, 5(6): 1–12, December 2009.
[5]. Sudipto Mukherjee, Trent E. Balius, and Robert C. Rizzo. Docking validation resources: Protein family and ligand flexibility experiments. Journal of Chemical Information and Modeling, 50(11): 1986–2000, October 2010.
[6]. Schrödinger, LLC. The PyMOL molecular graphics system, version 1.8, 2015.
For more information, please visit or follow the WeChat public account " Yin Fu Technology ".
Hot Plate Welding Machine,Hot Plate Plastic Welding Equipment,Servo Hot Plate Welding Machine,Hot Plate Welder For Oil Tank
Wuxi DIZO Ultrasonic Technology Company , https://www.dizosonic.com